The “growthstandards” R package provides methods for converting anthropometric measurements to or from z-scores and centiles using the WHO and INTERGROWTH growth standards. The WHO standard are used for infants, children, and young adults. INTERGROWTH standards are provided for fetal and newborn (including very preterm) calculations.
This document provides a walk-through of much of the package’s functionality, provided through examples and discussion. The package reference page has an exhaustive listing of all of the exported functions in the package along with detailed documentation about their parameters and usage.
Installation
The growthstandards package is not yet on CRAN but can be installed from a CRAN-like repository with the following:
options(repos = c(
CRAN = "http://cran.rstudio.com/",
deltarho = "http://packages.deltarho.org"))
install.packages("growthstandards")Once the package is is installed, you simply need to load it:
library(growthstandards)Growth Standards
A major feature of this package is a collection of utility functions for conveniently converting anthropometric measurements to z-scores or centiles (and converting z-scores / centiles to measurements) for three major growth standards:
- WHO growth standard (functions prefixed with
who_) - INTERGROWTH newborn size standard (functions prefixed with
igb_) - INTERGROWTH fetal growth standard (functions prefixed with
igfet_)
These growth standards have previously not necessarily been easy to access inside an R analysis. In some cases R code has been provided for making conversions with the growth standard but in a form that is difficult to embed and generalize (copying and pasting code that will be frequently used is messy will almost surely lead to errors). Some standards are provided only through published coefficients. The goal here is to put all the standards into a single package with a unified interface, while providing additional functionality like interpolation for regions where a standard’s provided tables are sparse.
The growth standard conversion methods have been painstakingly checked for accuracy through comparisons with the standards provided by the original sources. However, we advise caution and double checking against the original sources when results will impact decisions. Links to the original sources can be found in the sections for each standard below.
WHO growth standard
The WHO growth standard provides international standard distributions for several anthropometric measurements.
The data / methodology for the WHO growth standard functions in this package come from the WHO child growth standard and the 5-19 years growth reference.
WHO growth standard conversions are available for the following pairings of variables:
| x_var | y_var | span of x_var |
|---|---|---|
| agedays | wtkg (weight kg) | ~10 years (0 - 3682 days) |
| agedays | htcm (height cm) | ~19 years (0 - 6970 days) |
| agedays | bmi (BMI kg/m2) | ~19 years (0 - 6970 days) |
| agedays | hcircm (head circumference cm) | ~ 5 years (0 - 1856 days) |
| agedays | muaccm (mid upper-arm circumference cm) | ~ 5 years (0 - 1856 days) |
| agedays | ss (subscalpular skinfold) | ~ 5 years (0 - 1856 days) |
| agedays | tsftmm (triceps skinfold thickness mm) | ~ 5 years (0 - 1856 days) |
| htcm | wtkg | 120 cm |
Note that the WHO provides recumbent length for 0-2 years of age, after which the standing height is provided. Here we have merged the two into a single variable, htcm.
Also note that in almost every case the x variable is agedays - the age since birth in days. When the desired x variable is agedays, a simplified set of functions has been created to convert between z-scores or centiles with the following naming convention, for example, when working with wtkg:
-
who_wtkg2zscore(agedays, wtkg, sex = "Female"): for specifiedwtkgvalues at agesagedays, get the corresponding Female z-scores -
who_wtkg2centile(agedays, wtkg, sex = "Female"): for specifiedwtkgvalues at agesagedays, get the corresponding Female centiles -
who_centile2wtkg(agedays, p = 50, sex = "Female"): for given centiles (p=50, or median, as default) at agesagedays, get the correspondingwtkgvalues -
who_zscore2wtkg(agedays, z = 0, sex = "Female"): for given z-scores (z=0as default) at agesagedays, get the correspondingwtkgvalues
If you are working with htcm you can swap wtkg for htcm, etc.
As z-scores are related to centiles through a simple conversion, there is a bit of redundancy in having functions for both, but both are provided for convenience.
Examples
Here are some examples:
Get WHO median height vs. age for first-year females
Here we get the WHO median height vs. age for females weekly for the first year:
x <- seq(0, 365, by = 7)
med <- who_centile2htcm(x)
plot(x, med, xlab = "age in days", ylab = "median female height (cm)")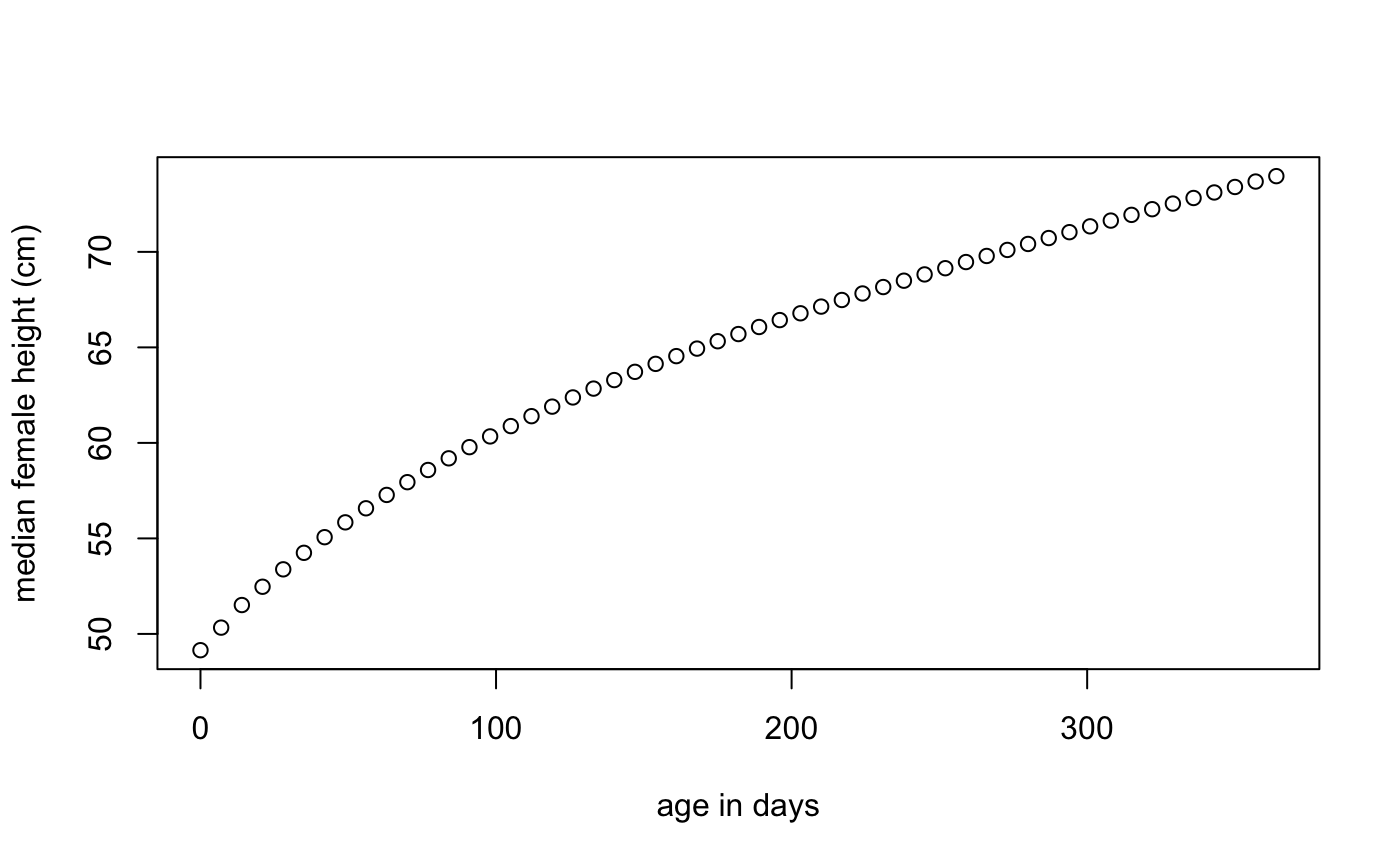
Here the defaults of sex="Female" and p=50 were used, giving us the median height for females at the specified ages.
Get WHO 75th centile height vs. age for first-year males
To get the WHO 75th centile weight value for age of males at these same time points, we can do the following:
q75 <- who_centile2htcm(x, p = 75, sex = "Male")Get the WHO centile for specific age and height
Suppose we want to know the WHO centile of a female child at 2 years of age (730.5 days) who is 90 cm tall:
who_htcm2centile(agedays = 730.5, htcm = 90)[1] 88.86932This girl’s height is at the 89th centile.
Getting values for combinations of parameters
We can send vectors of values into any of the functions and lookups in the appropriate tables will be made for each distinct case.
For example, suppose we want to plot the WHO 99th centile of weight for age of boys and girls for the first 4 years of life:
dat <- data.frame(
x = rep(seq(0, 1461, length = 100), 2),
sex = rep(c("Male", "Female"), each = 100))
dat$p99 <- who_centile2wtkg(dat$x, p = 99, sex = dat$sex)
library(lattice)
xyplot(kg2lb(p99) ~ days2years(x), groups = sex, data = dat,
ylab = "99th percentile weight (pounds) for males",
xlab = "age (years)", auto.key = TRUE)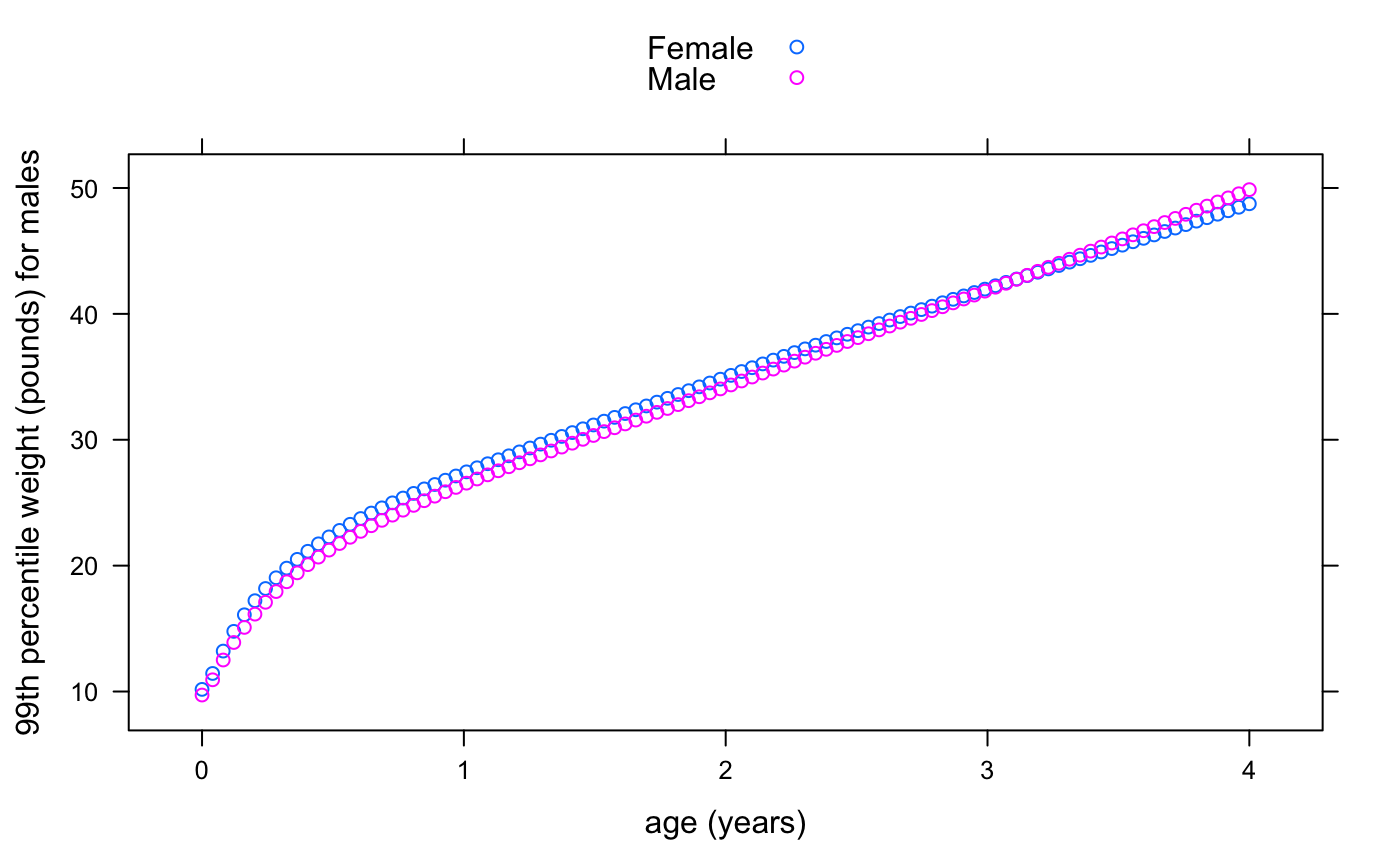
Here we are using a convenience function, kg2lb(), provided with the package to plot the weights in pounds.
Getting z-scores for combinations of parameters
Suppose we want to compute the height and weight-for-age z-scores (HAZ and WAZ) for each subject in the cpp data:
haz <- who_htcm2zscore(cpp$agedays, cpp$htcm, sex = cpp$sex)
waz <- who_wtkg2zscore(cpp$agedays, cpp$wtkg, sex = cpp$sex)Generic WHO functions
To utility functions described so far all for measurements against age. To deal with the case of height-for-weight WHO calculations, and also to provide a more generic interface to accessing the WHO growth standards, the following functions are also provided:
who_value2zscore(x, y, x_var = "agedays", y_var = "htcm", sex = "Female")who_value2centile(x, y, x_var = "agedays", y_var = "htcm", sex = "Female")who_centile2value(x, p = 50, x_var = "agedays", y_var = "htcm", sex = "Female")who_zscore2value(x, z = 0, y_var = "htcm", x_var = "agedays", sex = "Female")
Here both the x variable and y variable can be specified. For example, the following are equivalent:
who_htcm2centile(agedays = 730.5, htcm = 90)[1] 88.86932who_value2centile(x = 730.5, y = 90, x_var = "agedays", y_var = "htcm")[1] 88.86932But now we can do things like computing height-for-weight centiles:
who_value2centile(x = 90, y = 12, x_var = "htcm", y_var = "wtkg")[1] 27.61003INTERGROWTH newborn standard
The INTERGROWTH newborn standard provides standards for newborn weight, length, and head circumference by gestational age and sex from gestational age of 232 days to 300 days.
Functions for this standard have a similar naming structure as we saw for the WHO standards:
-
igb_lencm2zscore(gagebrth, lencm, sex = "Female"): for specifiedlencmvalues at gestational ages at birthgagebrth, get the corresponding Female z-scores -
igb_lencm2centile(gagebrth, lencm, sex = "Female"): for specifiedlencmvalues at gestational ages at birthgagebrth, get the corresponding Female centiles -
igb_centile2lencm(gagebrth, p = 50, sex = "Female"): for given centiles (p=50, or median, as default) at gestational ages at birthgagebrth, get the correspondinglencmvalues -
igb_zscore2lencm(gagebrth, z = 0, sex = "Female"): for given z-scores (z=0as default) at gestational ages at birthgagebrth, get the correspondinglencmvalues
The same functions are available for weight wtkg and head circumference hcircm.
Usage is also the same as in WHO.
Examples
Suppose we want to get the birth length z-scores for all subjects in our cpp data set:
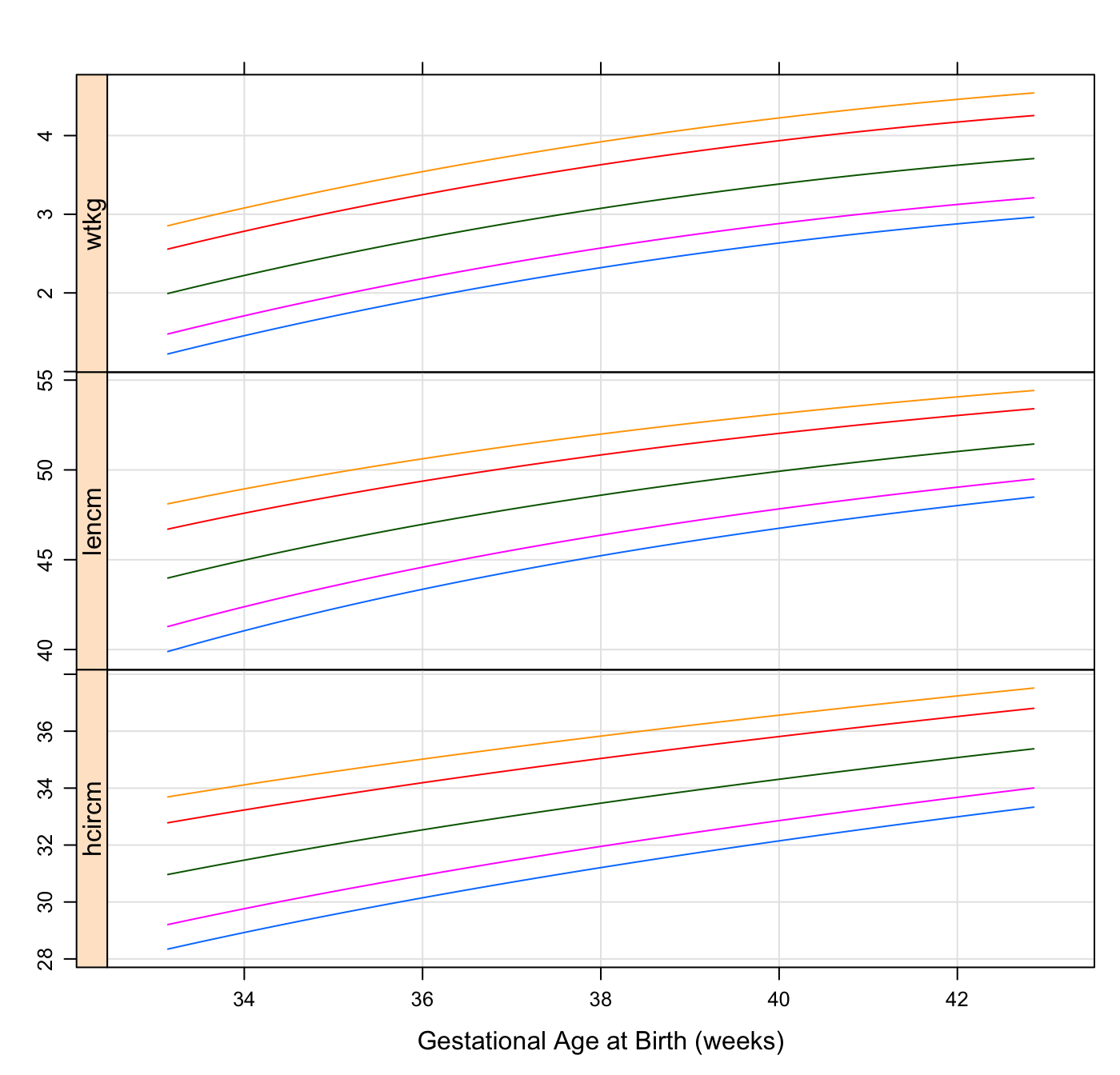
hbaz <- igb_lencm2zscore(cpp$gagebrth, cpp$birthlen, sex = cpp$sex)Also, we can make a quick and crude recreation of the boys charts seen here with this:
chartdat <- do.call(rbind, lapply(c(3, 10, 50, 90, 97), function(p) {
data.frame(p = p, gagebrth = 232:300,
make.groups(
wtkg = igb_centile2wtkg(232:300, p, sex = "Male"),
lencm = igb_centile2lencm(232:300, p, sex = "Male"),
hcircm = igb_centile2hcircm(232:300, p, sex = "Male")
))
}))
xyplot(data ~ gagebrth / 7 | which, groups = p, data = chartdat,
type = c("l", "g"),
strip = FALSE, strip.left = TRUE,
scales = list(y = list(relation = "free")),
layout = c(1, 3), as.table = TRUE,
xlab = "Gestational Age at Birth (weeks)", ylab = ""
)
INTERGROWTH prenatal standard
This package also has functions for dealing with the INTERGROWTH international standards for fetal growth based on serial ultrasound measurements. This data covers gestational ages from 7 to 40 weeks (98 to 280 days), for the following variables:
| variable | description |
|---|---|
| hccm | head circumference (cm) |
| bpdcm | biparietel diameter (cm) |
| ofdcm | occipito-frontal diameter (cm) |
| accm | abdominal circumference (cm) |
| flcm | femur length (cm) |
As with previous growth standard methods, a similar interface to this growth standard is provided with the following conventions for, for example, hccm:
-
igfet_flcm2zscore(gagedays, flcm, sex = "Female"): for specifiedflcmvalues at gestational agesgagedays, get the corresponding Female z-scores -
igfet_flcm2centile(gagedays, flcm, sex = "Female"): for specifiedflcmvalues at gestational agesgagedays, get the corresponding Female centiles -
igfet_centile2flcm(gagedays, p = 50, sex = "Female"): for given centiles (p=50, or median, as default) at gestational agesgagedays, get the correspondingflcmvalues -
igfet_zscore2flcm(gagedays, z = 0, sex = "Female"): for given z-scores (z=0as default) at gestational agesgagedays, get the correspondingflcmvalues
Usage is similar as the other growth standards. For example, to get the centile for child at 100 gestational days with an ultrasound head circumference measurement of 11cm:
igfet_hccm2centile(100, 11)[1] 93.39812Plot utilities
This package provides a few utility functions for making it easy to add any of the growth standards as an overlay on a plot. Methods are available for ggplot, lattice, and rbokeh. These functions have similar parameters to the growth standard lookup functions described above. The methods available are:
- ggplot2
geom_who()geom_igb()geom_igfet()- lattice
panel.who()panel.igb()panel.igfet()- rbokeh
ly_who()ly_igb()ly_igfet()
Some examples are below.
ggplot
Here is an example of using the utility function, geom_who(), with a plot of a CPP subject’s height vs. age, plotting the default of WHO bands at centiles 1, 5, 25, 50, 75, 95, 99:
library(ggplot2)
ggplot(data = subset(cpp, subjid == 8), aes(x = agedays, y = htcm)) +
geom_who(x_seq = seq(0, 2600, by = 10), y_var = "htcm") +
geom_point() +
theme_bw()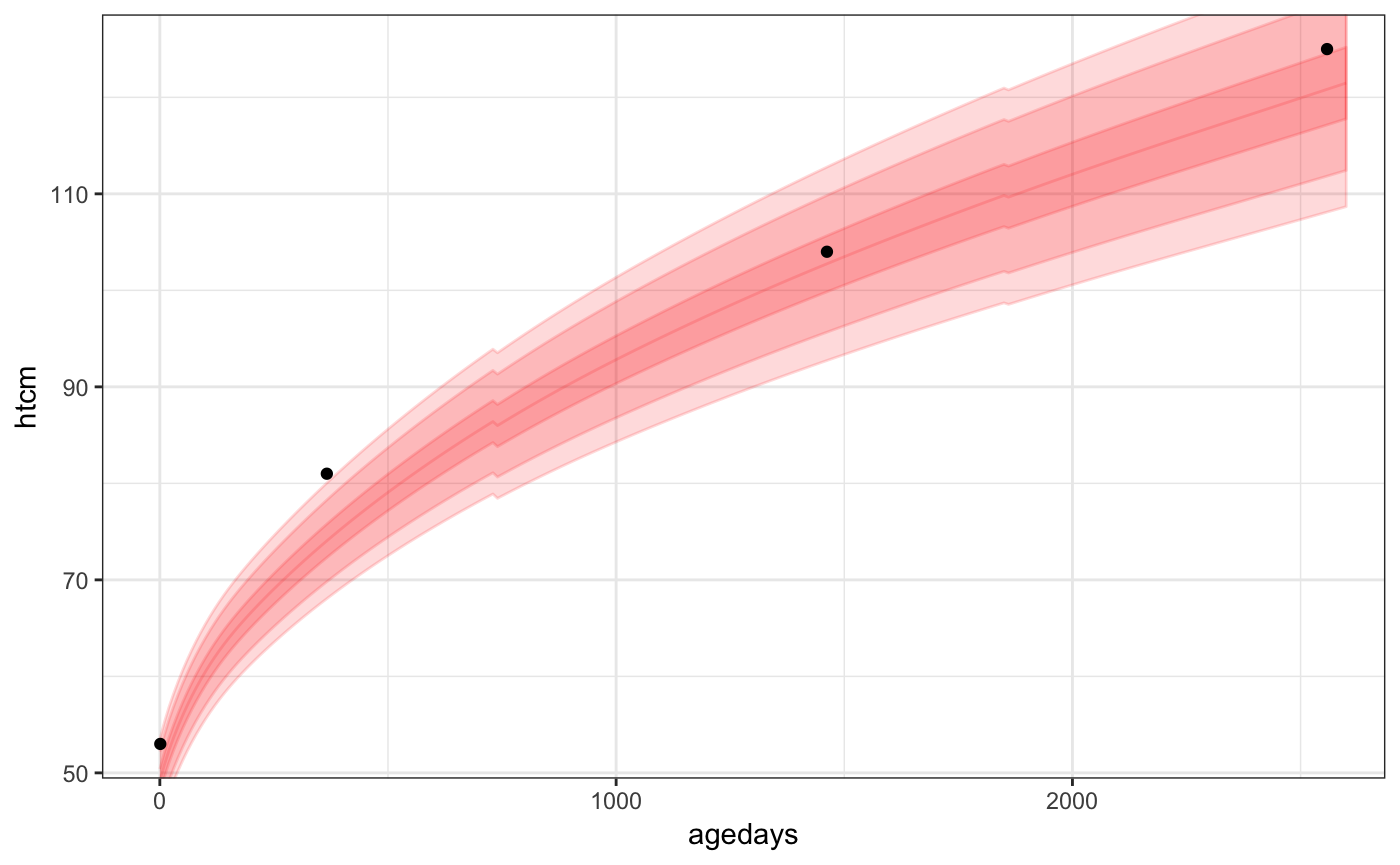
lattice
This example of uses panel.who() to superpose the WHO growth standard with a plot of a CPP subject’s height vs. age, with WHO bands at z-scores -3, -2, -1, 0, 1, 2, 3:
xyplot(wtkg ~ agedays, data = subset(cpp, subjid == 8),
panel = function(x, y, ...) {
panel.who(x = seq(0, 2600, by = 10),
sex = "Male", y_var = "wtkg", p = pnorm(-3:0) * 100)
panel.xyplot(x, y, ...)
},
ylim = c(0, 39),
col = "black")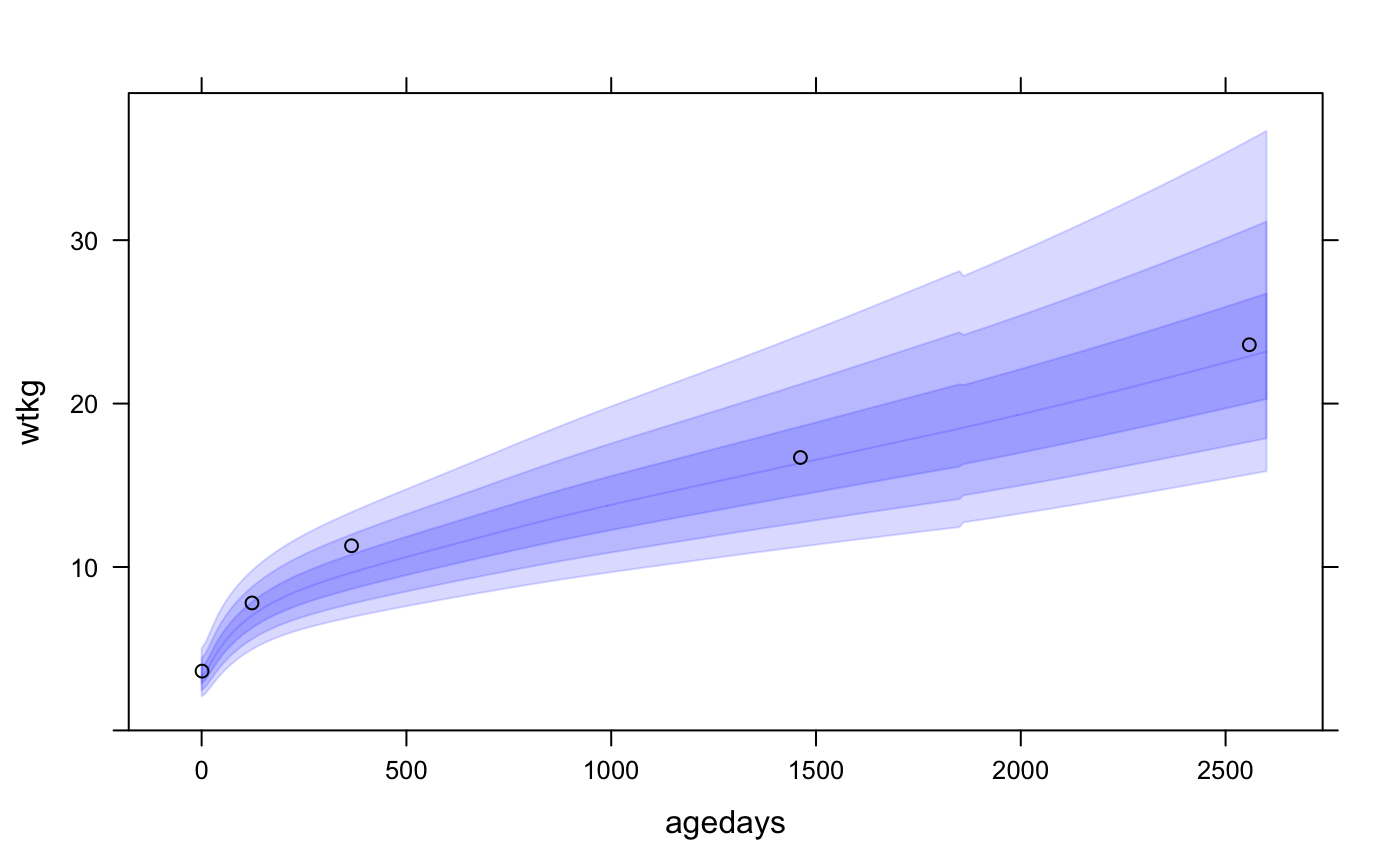
rbokeh
Here’s an example plotting the same plot as the lattice plot, transforming agedays to age in years and taking advantage of several of rbokeh’s interactive features, such as hover and zoom/pan.
library(rbokeh)
figure(xlab = "Age (years)", ylab = "wtkg") %>%
ly_who(x = seq(0, 2600, by = 10), y_var = "wtkg",
sex = "Male", x_trans = days2years) %>%
ly_points(days2years(agedays), wtkg,
data = subset(cpp, subjid == 8), col = "black",
hover = c(agedays, wtkg, lencm, htcm, bmi, geniq, sysbp, diabp))Warning in structure(x, class = unique(c("AsIs", oldClass(x)))): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
Consider 'structure(list(), *)' instead.
Warning in structure(x, class = unique(c("AsIs", oldClass(x)))): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
Consider 'structure(list(), *)' instead.
Warning in structure(x, class = unique(c("AsIs", oldClass(x)))): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
Consider 'structure(list(), *)' instead.
Warning in structure(x, class = unique(c("AsIs", oldClass(x)))): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
Consider 'structure(list(), *)' instead.
Warning in structure(x, class = unique(c("AsIs", oldClass(x)))): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
Consider 'structure(list(), *)' instead.
Warning in structure(x, class = unique(c("AsIs", oldClass(x)))): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
Consider 'structure(list(), *)' instead.
Warning in structure(x, class = unique(c("AsIs", oldClass(x)))): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
Consider 'structure(list(), *)' instead.
Warning in structure(x, class = unique(c("AsIs", oldClass(x)))): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
Consider 'structure(list(), *)' instead.
Warning in structure(x, class = unique(c("AsIs", oldClass(x)))): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
Consider 'structure(list(), *)' instead.
Warning in structure(x, class = unique(c("AsIs", oldClass(x)))): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
Consider 'structure(list(), *)' instead.
Warning in structure(x, class = unique(c("AsIs", oldClass(x)))): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
Consider 'structure(list(), *)' instead.Look at Male birth lengths superposed on the INTERGROWTH birth standard:
# first we need just 1 record per subject with subject-level data
cppsubj <- cpp[!duplicated(cpp$subjid),]
figure(xlab = "Gestational Age at Birth (days)", ylab = "Birth Length (cm)") %>%
ly_igb(gagebrth = 250:310, var = "lencm", sex = "Male") %>%
ly_points(jitter(gagebrth), birthlen, data = subset(cppsubj, sex == "Male"),
color = "black")Warning in structure(x, class = unique(c("AsIs", oldClass(x)))): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
Consider 'structure(list(), *)' instead.
Warning in structure(x, class = unique(c("AsIs", oldClass(x)))): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
Consider 'structure(list(), *)' instead.
Warning in structure(x, class = unique(c("AsIs", oldClass(x)))): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
Consider 'structure(list(), *)' instead.
Warning in structure(x, class = unique(c("AsIs", oldClass(x)))): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
Consider 'structure(list(), *)' instead.
Warning in structure(x, class = unique(c("AsIs", oldClass(x)))): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
Consider 'structure(list(), *)' instead.
Warning in structure(x, class = unique(c("AsIs", oldClass(x)))): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
Consider 'structure(list(), *)' instead.
Warning in structure(x, class = unique(c("AsIs", oldClass(x)))): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
Consider 'structure(list(), *)' instead.
Warning in structure(x, class = unique(c("AsIs", oldClass(x)))): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
Consider 'structure(list(), *)' instead.
Warning in structure(x, class = unique(c("AsIs", oldClass(x)))): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
Consider 'structure(list(), *)' instead.
Warning in structure(x, class = unique(c("AsIs", oldClass(x)))): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
Consider 'structure(list(), *)' instead.
Warning in structure(x, class = unique(c("AsIs", oldClass(x)))): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
Consider 'structure(list(), *)' instead.Plot INTERGROWTH prenatal growth standard bands at z=1,2,3 for prenatal head circumference
figure(xlab = "Gestational Age (days)",
ylab = "Head Circumference (cm)") %>%
ly_igfet(gagedays = 98:280, var = "hccm", p = pnorm(-3:0) * 100)Warning in structure(x, class = unique(c("AsIs", oldClass(x)))): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
Consider 'structure(list(), *)' instead.
Warning in structure(x, class = unique(c("AsIs", oldClass(x)))): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
Consider 'structure(list(), *)' instead.
Warning in structure(x, class = unique(c("AsIs", oldClass(x)))): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
Consider 'structure(list(), *)' instead.
Warning in structure(x, class = unique(c("AsIs", oldClass(x)))): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
Consider 'structure(list(), *)' instead.
Warning in structure(x, class = unique(c("AsIs", oldClass(x)))): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
Consider 'structure(list(), *)' instead.
Warning in structure(x, class = unique(c("AsIs", oldClass(x)))): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
Consider 'structure(list(), *)' instead.
Warning in structure(x, class = unique(c("AsIs", oldClass(x)))): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
Consider 'structure(list(), *)' instead.
Warning in structure(x, class = unique(c("AsIs", oldClass(x)))): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
Consider 'structure(list(), *)' instead.
Warning in structure(x, class = unique(c("AsIs", oldClass(x)))): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
Consider 'structure(list(), *)' instead.
Warning in structure(x, class = unique(c("AsIs", oldClass(x)))): Calling 'structure(NULL, *)' is deprecated, as NULL cannot have attributes.
Consider 'structure(list(), *)' instead.